Cytosplore Transcriptomics
Cytosplore Transcriptomics incorporates state-of-the art clustering and dimensionality reduction techniques for interactive exploration of cell types. One can visualize transcriptome-wide gene expression in combination with metadata of individual cells, perform differential analyses and statistics between manual selections of cells. Using GPU accelarated tSNE and HSNE Cytosplore Transcriptomics is scalable to large datasets having millions of cells, and can interactively produce low-dimensional visualizations of the data hierarchy providing different levels of details.
Cytosplore Transcriptomics is based on Cytosplore and provides many of the analysis tools presented in the previous Cytosplore publications and is available for download. Cytosplore Transcriptomics makes use of HDILib for fast tSNE and HSNE computations.
Team
LUMC
Development
LUMC
Computational Biology
LUMC
Computational Biology
TU Delft / LUMC
TU Delft
Project Lead Cytosplore
LUMC
PI LKEB
Cytosplore Transcriptomics is being developed by a team consisting of members from the Leiden Computational Biology Center, the Division of Image Processing at the Leiden University Medical Center, and the Computer Graphics and Visualization Group at the TU Delft
This work was partially supported by an NWO Gravitation grant:BRAINSCAPES: A Roadmap from Neurogenetics to Neurobiology (NWO: 024.004.012) and NWO AES grants 3DOMICS (NWO: 17126), VANPIRE (NWO: 12720) and Genes in Space (NWO: 12721).
Publications
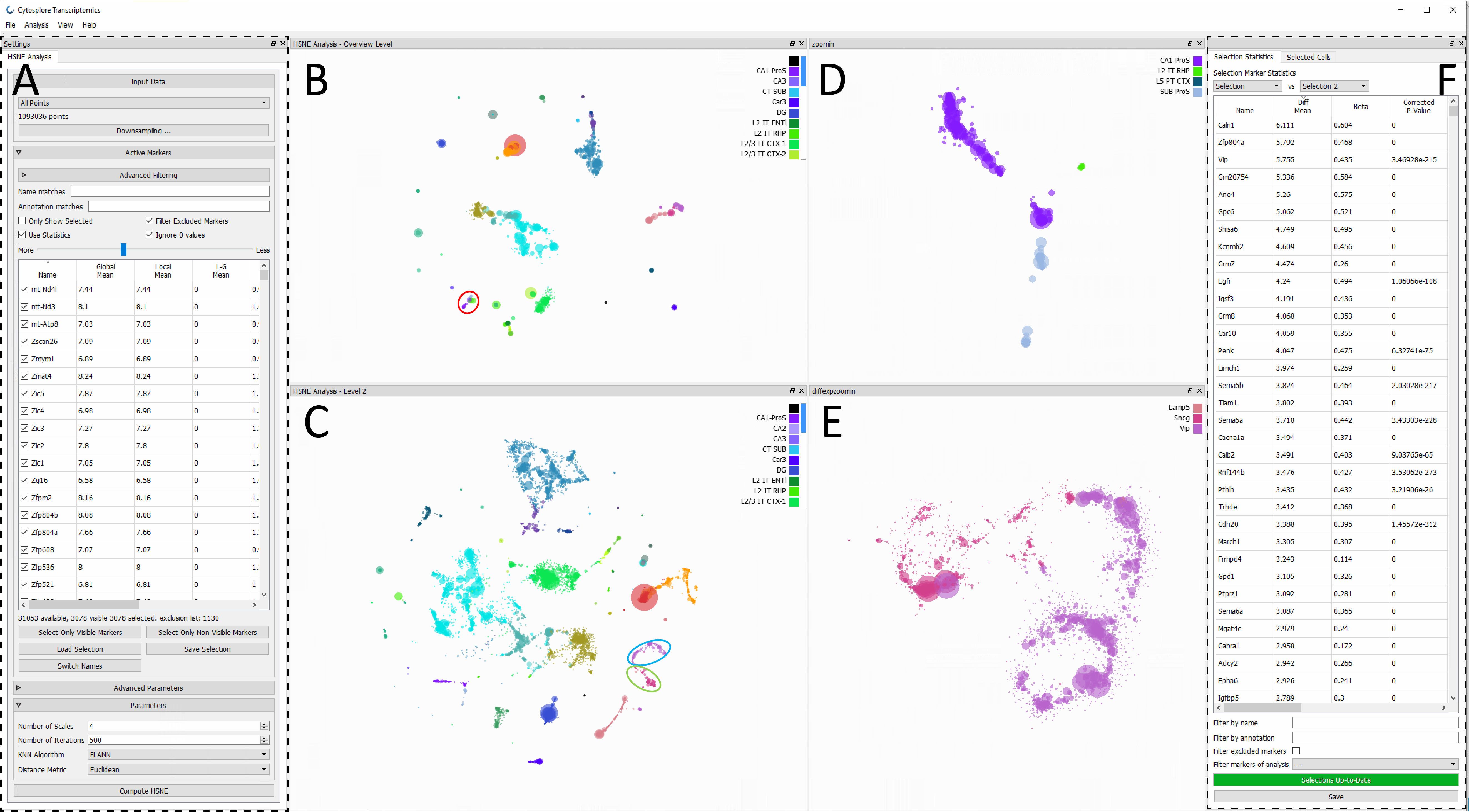
Cytosplore-Transcriptomics: a scalable inter-active framework for single-cell RNA sequenc-ing data analysis , BioRxiv, 2020.
Abstract: The ever-increasing number of analyzed cells in Single-cell RNA sequencing (scRNA-seq) experiments imposes several challenges on the data analysis. Current analysis methods lack scalability to large datasets hampering interactive visual exploration of the data. We present Cytosplore-Transcriptomics, a framework to analyze scRNA-seq data, including data preprocessing, visualization and downstream analysis. At its core, it uses a hierarchical, manifold preserving representation of the data that allows the inspection and annotation of scRNA-seq data at different levels of detail. Consequently, Cytosplore-Transcriptomics provides interactive analysis of the data using low-dimensional visualizations that scales to millions of cells. ...
Get Cytosplore Transcriptomics
Disclaimer: Cytosplore Transcriptomics is a research project between TU Delft and Leiden University Medical Center. We do our best to provide support but cannot guarantee it. If you have any suggestions, problems, want to share your success stories or papers published using Cytosplore Transcriptomics, we would love to get feedback! Please do not hesitate to get in touch.
If you use Cytosplore Transcriptomics within the scope of a scientific article you must cite the original publications:
Abdelaal et al. Cytosplore-Transcriptomics: a scalable inter-active framework for single-cell RNA sequenc-ing data analysis, BioRxiv, 2020.
For other operating systems or older versions please see the release archive.